Michigan State University plant biologists have made a discovery that could help turn a natural kill switch in plant cells into a “life switch” that helps crops better survive the challenges presented by climate change.

At its core, though, this is a fundamental finding, shared in the journal Nature Plants, that has implications across biology for how organisms respond to stress linked to overproduction of proteins by the cell.
“Life depends on the activity of an organelle called the endoplasmic reticulum, or ER,” said Federica Brandizzi, the leader of the laboratory that published the new finding.
Brandizzi is an MSU Distinguished Professor and MSU Research Foundation Professor in the Department of Plant Biology and the MSU-Department of Energy Plant Research Laboratory.
“The ER produces essential biomolecules, including lipids and a third of the proteins used by cells. It also facilitates cells’ communication with the external environment,” Brandizzi said. “Certain physiological and stress situations can lead to a failure of the biosynthetic ability of this organelle, a situation known as ER stress, which can be lethal.”
“What we’ve found is a specific pathway and new regulators not known to be involved with ER stress responses before,” said the study’s lead author, Dae Kwan Ko, an assistant professor in the Brandizzi lab at MSU. “This discovery opens new doors and new directions in research.”
Stress and a cellular kill switch
Cells in every eukaryotic organism — plants, fungi and animals, including humans — have numerous self-destruct mechanisms they can activate when they’re under unfavorable environmental conditions.
Cells sacrificing themselves can help preserve the health of the larger organism under certain conditions — by stopping the spread of a disease, for example. Under other conditions, though, death at the cellular level can cascade into harm, illness and even death for the organism.
“By understanding these biomolecular self-destruct mechanisms in cells, researchers could devise tactics to avoid or delay when a cell activates them in response to certain stressors,” said Ko.

Unfortunately, these mechanisms are largely mysterious and highly complicated. Fortunately, Ko and his co-authors — also members of Brandizzi’s lab — are skilled at simplifying. Joining Ko and Brandizzi on the project were Joo Yong Kim, a postdoctoral research associate, and Ethan Thibault, a doctoral student.
Ko said that there’s an interplay of genes and protein activity that relay stress signals to the cell’s command center, or nucleus. When a certain signaling pathway is activated, “it’s like a switch between life and death during a stress response,” said Ko.
Ko and his colleagues devised experiments that identified proteins regulating one of these pathways, along with the associated genes.
“In this paper, we tried to identify the regulators of one signaling pathway,” Ko said. “There’s really not much known about which proteins do what, where and when. We want to understand what they’re doing in time and space.”
To do this, Ko and the team focused on illuminating a single mechanism or pathway in ER stress.
Putting a model organism to work
The ER is an organelle that cells in all eukaryotes use to fold proteins, among other things. Under normal conditions, a cell’s need to have proteins folded is balanced by the ER’s capacity to fold them. It’s like driving on a freeway with light traffic, Ko said.
But when cells grow or undergo certain stresses, including attack by pathogens, the demand for protein folding outpaces capacity. This leads to a traffic jam of unfolded proteins. That’s ER stress and, when it gets too severe, it can be fatal.
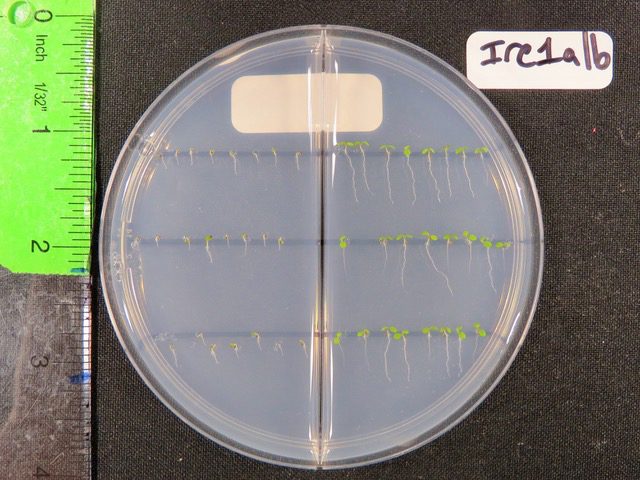
To zero in on one of the pathways that cells use to decide where that tipping point is, the team turned to a model organism, a plant known as Arabidopsis thaliana or thale cress. Using a model like thale cress, scientists can also start identifying genes and traits that are shared, or conserved, in other species.
“These processes are highly conserved, not just in plants, but in animals and all eukaryotes,” Ko said. “Studying these processes in a model system like Arabidopsis has the advantage of letting us carry out research fast using ample genomics resources.”
The team grew “regular” Arabidopsis plants along with other lineages that had random genetic mutations. All told, the team created plants that encompassed thousands of genetic changes.
The researchers then watched how the plants matured after being exposed to a compound that inhibits protein folding. That is, the researchers essentially kickstarted the traffic jam on the ER highway.
While normal plants could withstand this stress, a particular mutant missing a protein known as IRE1 — short for “Inositol Requiring Enzyme 1” — could not. But further mutations to this mutant could return a plant’s ER stress response closer to normal.
“We have this mutant plant that is supposed to be sick under ER stress because it does not have a protein necessary for ER stress responses,” Ko said. “But by mutagenizing the mutant, we found another mutant that reverts the sickness.”
In particular, this more resilient mutant lost an additional protein named PIR1 (short for “Phosphatase type 2CA Interacting Ring finger protein 1”). Working with the Research Technology Support Facility Genomics Core and the Mass Spectrometry and Metabolomics Core facilities at MSU, the researchers also discovered the associated genetics and molecular signaling that determined a cell’s fate in ER stress conditions.

Though this is one pathway in one plant, there’s power in that plant being a model organism like Arabidopsis. The team’s methodology, for example, could be used to look for other important ER stress pathways found in other eukaryotes, such as humans.
And, although PIR1 is only found in plants, it’s found in hundreds of species, including crops like soybeans.
“So, you can start thinking about manipulating the gene activity in plants like soybeans to make them more resilient toward climate change,” Ko said.
“Although PIR1 is not a conserved protein outside the plant kingdom, it is likely that nonplant species use mechanisms similar to those guided by PIR1 to control life-or-death outcomes,” Brandizzi said. “Therefore, our research results can potentially influence research on ER stress management also in nonplant species.”
For Ko, though, there are many other interesting avenues still left to explore in Arabidopsis thaliana itself. For one, plant roots have about a dozen different cell types and understanding if and how this signaling pathway works differently in different cells could have implications for cell health.
“Because the ER is a biosynthetic factory in the cell, understanding how we can manage protein production in the ER has important implications for improving quality of plant biomass and our ability to use plants as large-scale bioreactors for the production of recombinant pharmaceutical proteins, such as antibodies and vaccines,” Brandizzi said.
So, this discovery is a bit like the roots of a germinating plant: Its extent is sure to grow broader and deeper.
This story was originally posted at the College of Natural Science
Is your organization interested in sponsoring MSU plant biology research? Click Here

